FocusStack and StimServer
New, open-source toolboxes for visual stimulation and analysis of two-photon imaging data
1st December, 2014
Figure 1: Overview of an experiment setup.
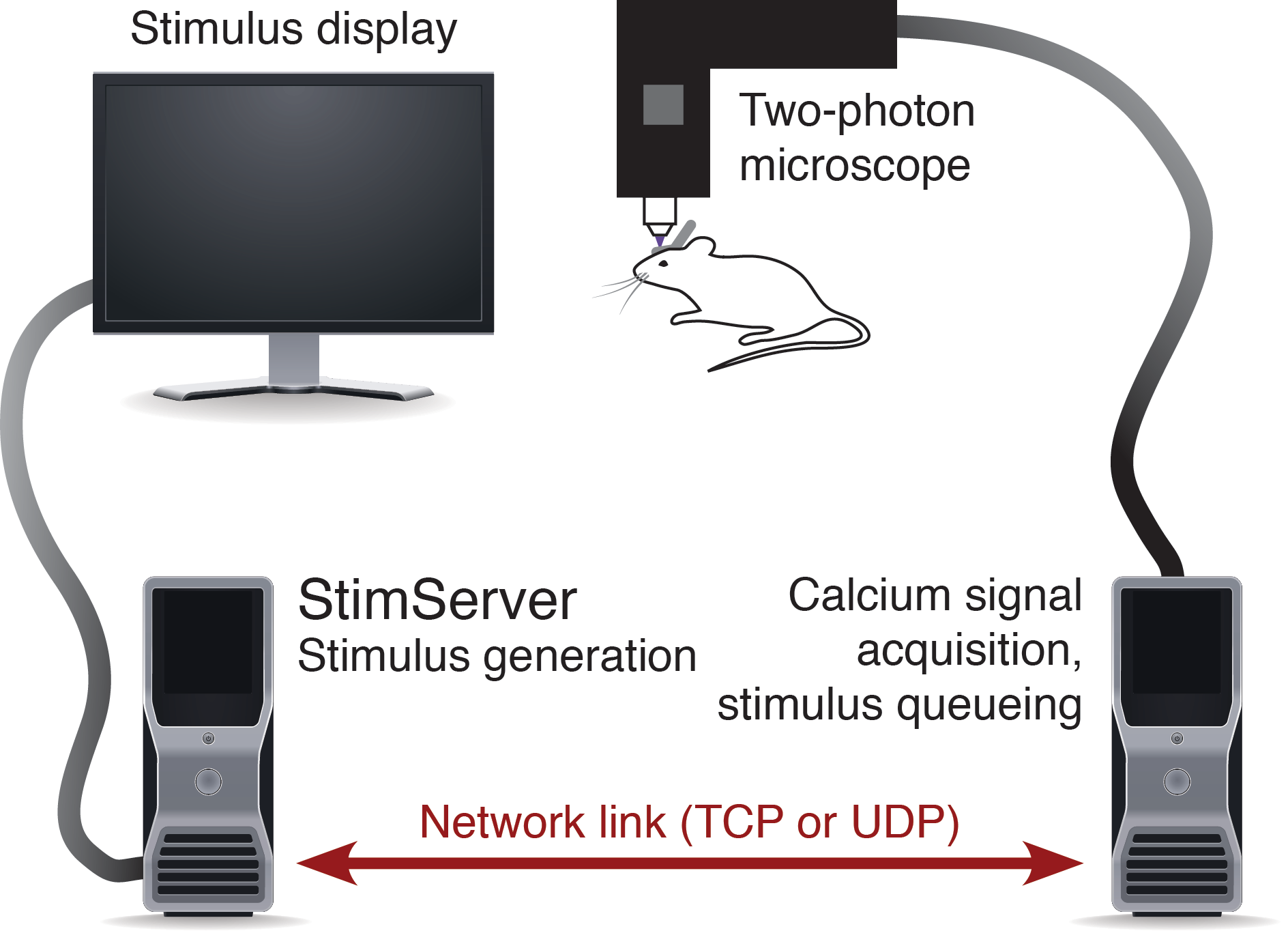
Figure 1: Overview of an experiment setup.
Figure 2: Importing and using raw two-photon imaging stacks.

Figure 2: Importing and using raw two-photon imaging stacks.
Figure 3: Frame meta-data
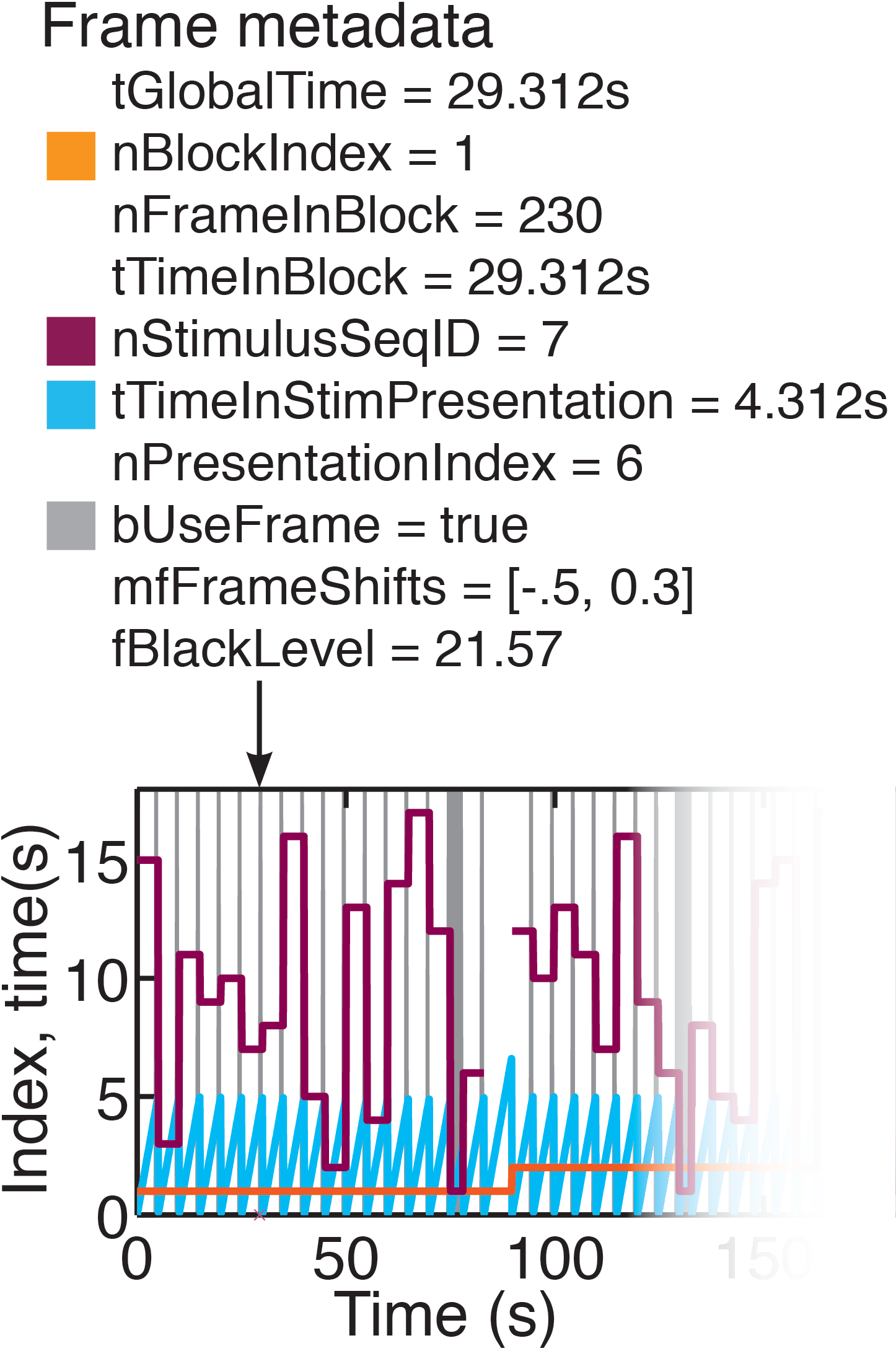
Figure 3: Frame meta-data
Figure 4: Extracting orientation-tuned responses
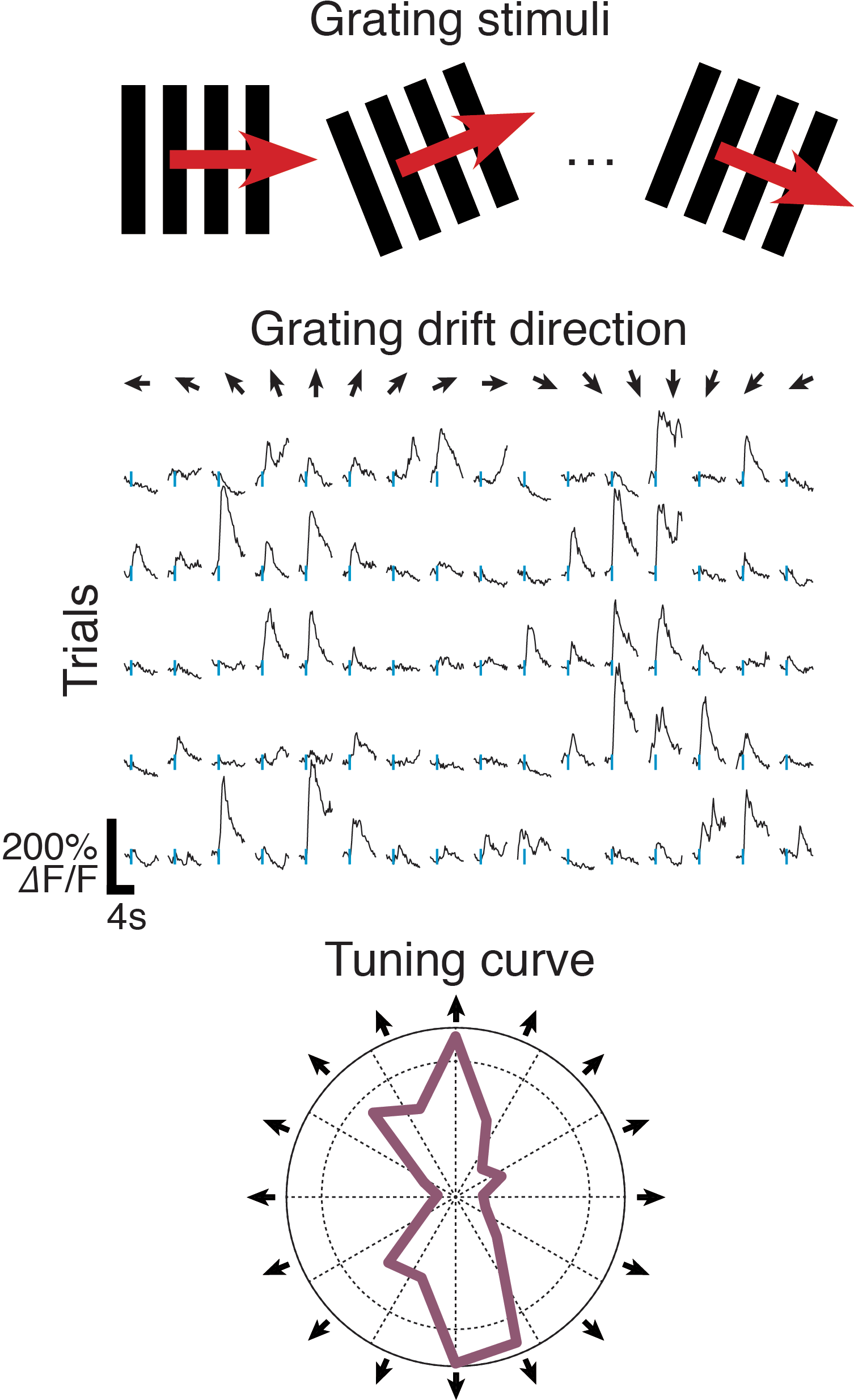
Figure 4: Extracting orientation-tuned responses
Two-photon calcium imaging of neuronal responses is an increasingly accessible technology for probing population responses in cortex at single cell resolution, and with reasonable and improving temporal resolution. However, analysis of two-photon data is usually performed using ad-hoc solutions. To date, no publicly available software exists for straightforward analysis of stimulus-triggered two-photon imaging experiments, while incorporating information about stimulus triggers. In addition, the increasing data rates of two-photon acquisition systems imply increasing cost of computing hardware required for in-memory analysis.
FocusStack is a new, open-source, Matlab-based two-photon analysis toolchain designed to process large two-photon imaging stacks with only a small memory footprint. This makes analysis possible on comparatively cheap consumer hardware. FocusStack is designed to integrate with StimServer, a new open-source Matlab-based server for visual stimulus generation and presentation which can be controlled remotely over TCP or UDP network links.
Accessing the toolboxes
Both toolboxes are available as open source repositories:
FocusStack: Wiki; GIT repository; 20141118 release (.zip)
StimServer: Wiki; GIT repository;
20141118 release (.zip)
Getting started with FocusStack
- Download the toolbox, either as a revision-controlled source repository using your favourite GIT utility, or download the zipped source code release linked above. Add the source directory to your Matlab path.
- (Optional) Matlab should be configured so that the
mexcompiler can successfully compile code. This allows the accelerated low-level compiled functions within the toolboxes to be used. - The paper describing
FocusStackandStimServerserves as a useful introduction to both toolboxes and their usage. The paper has been accepted for publication in Frontiers in NeuroInformatics. - Example analysis code, used to generate the figures in the
FocusStackpaper, is available to get you started with the toolbox. Download and unzip the example code (.zip). You will also need the example two-photon imaging data set (.zip) analysed in the paper.
Working with FocusStack
The design goal of FocusStack was to provide a simple, extensible toolchain to assist experiments using two-photon calcium imaging of neuronal responses; accessible to those with little programming experience, but powerful enough to automate most low-level analysis of calcium response stacks.
Due to the real-time requirements of both visual stimulation and acquisition of two-photon imaging data, these tasks are usually performed on separate dedicated computing systems. StimServer is controlled over a TCP or UDP network link, to trigger stimulus presentation and sequencing (Figure 1). Two-photon acquisition occurs using the software appropriate for the experimental equipment used, and stores the resulting imaging stacks as binary data files on disk. Ideally, meta-data about the stack — stimulus identity and random sequencing, stack resolution, information about the acquisition system, etc. — are stored with the stack data files in a file header or a “side-car” meta-data file.
Binary stack data files are analysed in Matlab, by using FocusStack to map several stack files to a single FocusStack object (Figure 2). This object appears as a simple Matlab tensor, with frames, channels and single pixels accessed using standard Matlab referencing. Since FocusStack objects can be accessed as Matlab tensors, many existing Matlab analysis functions that expect tensors can seamlessly be passed FocusStack objects without modification.
However, FocusStack objects are aware of stimulus timing and sequencing, provide services for stack alignment, provide support for assigning baseline fluorescence distributions, and have simple helper functions to perform derandomization and segmentation of stack data. Although a "FocusStack" object can be accessed just like a Matlab tensor, each frame and pixel in the stack has many items of meta-data associated with it (Figure 3). Stack-global information such as the stack resolution, or stimulus-specific meta data such as the stimulus presented during the acquisition of a given frame, is accessed using the FrameStimulusInfo method.
Calcium trace extraction and stimulus derandomisation
In any good experiment design, stimulus presentation order is randomized. During analysis of the acquired time series data, stimulus segmentation and derandomization therefore becomes an important but fiddly task. Our solution is to store the stimulus presentation order with the stack, along with information about stimulus duration, “blank” stimuli, and periods of stimulus presentation during which analysis of the calcium signals should be performed (see Figure 3).
Extracting calcium response time-series from a FocusStack object is accomplished using the ExtractRegionResponses function. This workhorse function transparently performs stimulus derandomization, simultaneously averages and extracts responses from a number of arbitrary ROIs in the stack, segments the stack into single-trial per-neuron traces and returns estimated responses for each stimulus and each trial.
The following code illustrates how complete calcium traces, as well as derandomised single-trial stimulus responses, can be extracted from experimental data in five lines of Matlab code. Note that information concerning stimulus presentation sequence is usually loaded along with the raw imaging data, in which assigning cvnSequenceIDs manually is unnecessary.
% - Create a FocusStack object
fs = FocusStack({'block1.bin', 'block2.bin'});
% - Assign stimulus presentation order
% (usually loaded from stack data file)
fs.cvnSequenceIDs = {[2 3 1 4], [4 2 3 1]};
% - Assign stimulus timing information
fs.vtStimulusDurations = [4 4 4 4];
fs.mtStimulusUseTimes = [0 4; 2 4; 2 4; 2 4];
% - Segment, derandomize and extract
% calcium responses and traces
[vfBlankStds, mfStimMeanResponses, mfStimStds, ...
mfRegionTraces, tfTrialResponses, tnFramesInSample, ...
cvfTrialTraces, mfRawRegionTraces] = ...
ExtractRegionResponses(fs, sRegions, nBlankStimID);
Analysis example — direction tuning
Figure 4 illustrates the extraction of orientation and direction tuning from multiple-trial calcium responses, evoked by drifting high-contrast grating stimuli presented to an anaesthetised mouse (using StimServer). This analysis is included in the ???example code linked above.
Publications
This work was published in Frontiers in Neuroinformatics: DR Muir and BM Kampa. 2015. FocusStack and StimServer: A new open source MATLAB toolchain for visual stimulation and analysis of two-photon calcium neuronal imaging data, Frontiers in Neuroinformatics 8 85. Please cite this publication in lieu of thanks, if you use either toolbox.
Acknowledgements
The authors would like to express effusive thanks to M. Roth for acquiring the experimental data analysed here, and for making that data available for download. We gratefully acknowledge the contributions of the users of FocusStack and
Funding
This work was supported by the Novartis Foundation (grant to DRM), Velux Stifting (grant to DRM), the Swiss National Science Foundation (Grant Nr. 31-120480 to BMK), and the EU-FP7 program (BrainScales project 269921 to BMK).